Macromolecular complexes and Membrane contact sites
We are interested in studying the molecular mechanisms leading to neurodegeneration. In particular, the role of disease-related proteins on organelle’s physiology and functioning is explored by combining cellular and animal models. Changes in protein subcellular localization and in the connectivity between organelles are increasingly linked to a growing number of diseases, being their impairment an early event in the disease onset. We have developed genetically encoded split-fluorescent proteins (FPs) based reporters to follow in vitro and in vivo these cellular processes.
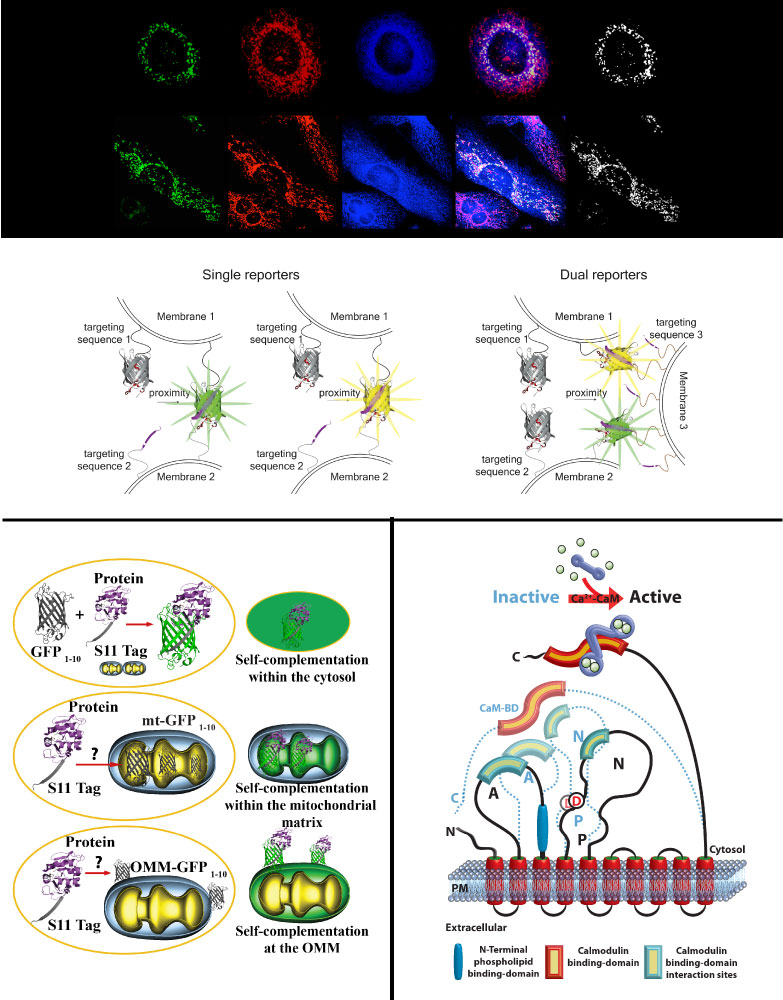
Constant bidirectional exchange of nutrients, metabolites, ions, and lipids, as well as protein function and localization at the interface between defined organelles kept in close apposition by defined factors or protein complexes is required to integrate them within specific cellular pathways thus guaranteeing cell fitness. These hubs called Membrane Contact Sites (MCS) are therefore crucial, and any deviation is linked to disease conditions such as neurodegeneration, cancer, or diabetes. We have generated the first genetically encoded reporter for in vitro and in vivo monitoring changes in the ER-mitochondria interface, and named it split-GFP-based contact site sensor (SPLICS). The SPLICS family was expanded to explore additional relevant contact sites such as ER-Plasma membrane, ER-peroxisomes, nucleus-mitochondria, mitochondria-peroxisomes or mitochondria-lysosomes. Further optimization allowed for the simultaneous detection of contact sites occurring at different membrane interfaces. The design of improved split-FP based reporters, the development of SPLICS reporters for novel disease-related contact sites and exploration of their behavior in living cells and in model organisms is our focus.
Multiple proteins linked to neurodegenerative diseases have multiple localizations associated to different functions. How and when shuttling among different subcellular compartments occur is still poorly defined. We have established split-GFP based reporters to detect protein translocation within defined subcellular compartments upon different cellular stimuli paving the way to the possibility to perform high-throughput screening.
A second topic of the research group is the structure-function relationship of the Plasma Membrane Calcium Pumps (PMCA) and the role of Ataxia-linked mutations of the neuronal isoforms. We have identified mutations of the PMCA in patients with Cerebellar Ataxia and the aim of the project is to study the effect of novel mutations of isoform 2 and 3 of the pump on its calcium pumping activity. We are also interested in achieving stuctural insights of the pump and other disease relevant macromolecular complexes purified from human erythrocytes.
People
Faculty
- Tito CALI' - Associate Professor
Lab Member
- Lucia Barazzuol - Post-doc
- Tetiana Tykhonenko - Post-doc
- Adamantia Denigiannopoulou - PhD student
Five recent publications
- Vallese, Francesca; Catoni, Cristina et al. An expanded palette of improved SPLICS reporters detects multiple organelle contacts in vitro and in vivo. Nature Communications, 11, 1, 6069. 2020
- Calì, Tito; Brini, Marisa; Quantification of organelle contact sites by split-GFP-based contact site sensors (SPLICS) in living cells. Nature Protocols, 16, 11, 5287-5308. 2021
- Cieri, Domenico; Vicario, Mattia; et al. SPLICS: a split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death & Differentiation, 25, 6, 1131-1145. 2018
- Vicario, Mattia; Cieri, Domenico et al. A split-GFP tool reveals differences in the sub-mitochondrial distribution of wt and mutant alpha-synuclein. Cell death & disease, 10, 11, 857. 2019
- Vallese, Francesca et al. The ataxia-linked E1081Q mutation affects the sub-plasma membrane Ca2+-microdomains by tuning PMCA3 activity. Cell Death & Disease, 13, 10, 855. 2022




